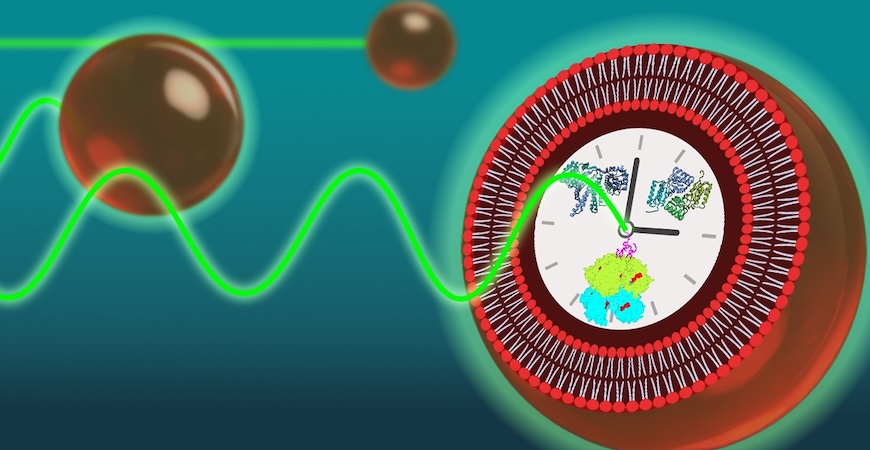
A team of UC Merced researchers has shown that tiny artificial cells can accurately keep time, mimicking the daily rhythms found in living organisms. Their findings shed light on how biological clocks stay on schedule despite the inherent molecular noise inside cells.
The study, recently published in Nature Communications, was led by bioengineering Professor Anand Bala Subramaniam and chemistry and biochemistry Professor Andy LiWang. The first author, Alexander Zhang Tu Li, earned his Ph.D. in Subramaniam’s lab.
Biological clocks — also known as circadian rhythms — govern 24-hour cycles that regulate sleep, metabolism and other vital processes. To explore the mechanisms behind the circadian rhythms of cyanobacteria, the researchers reconstructed the clockwork in simplified, cell-like structures called vesicles. These vesicles were loaded with core clock proteins, one of which was tagged with a fluorescent marker.
The artificial cells glowed in a regular 24-hour rhythm for at least four days. However, when the number of clock proteins was reduced or the vesicles were made smaller, the rhythmic glow stopped. The loss of rhythm followed a reproducible pattern.
To explain these findings, the team built a computational model. The model revealed that clocks become more robust with higher concentrations of clock proteins, allowing thousands of vesicles to keep time reliably — even when protein amounts vary slightly between vesicles.
The model also suggested another component of the natural circadian system — responsible for turning genes on and off — does not play a major role in maintaining individual clocks but is essential for synchronizing clock timing across a population.
The researchers also noted that some clock proteins tend to stick to the walls of the vesicles, meaning a high total protein count is necessary to maintain proper function.
“This study shows that we can dissect and understand the core principles of biological timekeeping using simplified, synthetic systems,” Subramaniam said.
The work led by Subramaniam and LiWang advances the methodology for studying biological clocks, said Mingxu Fang, a microbiology professor at Ohio State University and an expert in circadian clocks.
“The cyanobacterial circadian clock relies on slow biochemical reactions that are inherently noisy, and it has been proposed that high clock protein numbers are needed to buffer this noise,” Fang said. “This new study introduces a method to observe reconstituted clock reactions within size-adjustable vesicles that mimic cellular dimensions. This powerful tool enables direct testing of how and why organisms with different cell sizes may adopt distinct timing strategies, thereby deepening our understanding of biological timekeeping mechanisms across life forms.”
Subramaniam is a faculty member in the Department of Bioengineering and an affiliate of the Health Sciences Research Institute (HSRI). LiWang is a faculty member in the Department of Chemistry and Biochemistry, also affiliated with HSRI. He is a fellow of the American Academy of Microbiology and the 2025 recipient of the Dorothy Crowfoot Hodgkin Award from The Protein Society.
The work was supported by Subramaniam’s National Science Foundation CAREER award from the Division of Materials Research and by grants from the National Institutes of Health and Army Research Office awarded to LiWang. LiWang was supported by a fellowship from the NSF CREST Center for Cellular and Biomolecular Machines at UC Merced.



